run_2871_s_6_withindex_sequence.txt_ACGCGGG.fq.gz

 |
GTAC Summary of Services: |
| Computational Analysis: Demultiplexing Pipeline |
|
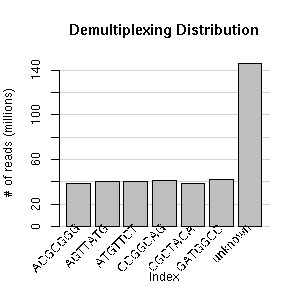
Project Details Elgin Lab Run: HiSeq 2871_6 Analyzed: 01/02/2019 Seq data expires: Analysis data expires: Run notes: |
1 Lane(s) single reads: 1 X 50 nts |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Deliverables:
If you have access to the HTCF cluster, the data is also located here: /scratch/gtac/GTAC_RUN_DATA/Elgin_2871_6
Raw sequence reads:
Files generated by the Illumina sequencer for single reads. (15-25G each -fastq format)
Any publications resulting from the attached data should include the following acknowledgment: "This publication was made possible in part by Grant Number UL1 RR024992 from the NIH-National Center for Research Resources (NCRR)."